Conditions for storing, freezing, and thawing biological molecules have become important topics of research in the biotechnology industry, as large biomolecules make up an large percentage of new reagents.
The following factors have been studied and shown to have major effects on biomolecule stability and activity: the rate of cooling during the freezing process, the storage temperature, the concentration at which the biomolecule is stored, the length of storage time, the pH, the presence of specific buffers and ions, the inclusion of cryoprotectants, lyophilization (freeze-drying), and numerous other factors [1–6].
However, some researchers still follow decades-old laboratory protocols for handling biomolecules without verifying their relevance to the compounds under study. For example, in many labs, it is thought essential to aliquot all protein solutions for single use, then store the aliquots at low temperatures (e.g., –80 or –20°C). Although this practice is intended to maintain protein quality and prevent contamination, it can lead to inaccurate concentrations used in experiments, as small aliquots are prone to evaporation. It can also increase denaturation of some proteins, since some protein molecules can adhere to plastic surfaces and denature [7].
Once proteins are frozen—whether as small aliquots or not—many researchers consider it necessary to thaw them on ice. Their reasoning is that this slow process is assumed to be gentle and unlikely to cause protein denaturation. However, rapid thawing has been shown to result in much better recovery of enzymatic activity compared to slow thawing for certain enzymes of mammalian and yeast origin [8].
Interestingly, different proteins can respond in opposite ways to the same freezing, storage, and thawing conditions. For example, lactate dehydrogenase is most stable when frozen slowly, while myoglobin is least stable when treated this way [5].
Utilizing the optimal freezing, storage, and thawing conditions for various biomolecules routinely used in research laboratories is important. Thus, IDT scientists have undertaken a study to determine optimal conditions for handling Alt-R CRISPR reagents. We have specifically studied length of time of storage at different temperatures, effects of different thawing temperatures, effects of varying the storage buffers, and effects of many freeze-thaw cycles.
In addition, we also investigated whether Cas nucleases can be pre-complexed with guide RNA (RNP complex) and then stored for an extended period of time before use. We found that all the Alt-R CRISPR reagents are remarkably resilient to a variety of conditions. Here, we offer guidelines for how best to handle these reagents.
Time in storage at various temperatures and concentrations
We have found that all Alt-R CRISPR reagents, when stored under the appropriate conditions, are stable at least 1 year. We recommend storing Alt-R CRISPR nucleases, guide RNA (crRNAs, tracrRNA, and sgRNA), and HDR Enhancer* at –20°C or –80°C. However, we also provide information on the behavior of the reagents during storage under other conditions (see Table 1 or download the PDF).
Table 1. Summary of CRISPR reagent stability**.
| Storage temperature | |||||
|---|---|---|---|---|---|
| –80ºC | –20ºC | 4ºC | 23ºC | ||
| Alt-R CRISPR nucleases |
| Stable up to 2 years in stock storage buffer | Stable up to 2 years in stock storage buffer | Stable up to 2 months in stock storage buffer | Stable up to 3 days in stock storage buffer |
| Stable up to 1 year in stock storage buffer | Stable up to 1 year in stock storage buffer | |||
| Alt-R CRISPR ribonucleoprotein complexes |
| Stable up to 2 years | Stable up to 2 years | Stable up to 2 months | Stable up to 3 days |
| Stable up to 1 year | Stable up to 1 year | |||
| Alt-R CRISPR guide RNAs |
| Stable up to 18 months:
| Stable up to 18 months:
|
|
|
| Stable up to 1 year:
| Stable up to 1 year:
|
|
| |
| CRISPR-Cas9 tracrRNA | Stable up to 2 years:
| Stable up to 2 years:
|
| ||
| Labeled tracrRNA, ATTO™ 550 | Stable up to 2 years:
|
|
| ||
| gRNA 2-part complex, made of CRISPR-Cas9 crRNA plus CRISPR-Cas9 tracrRNA | Stable up to 2 years in IDTE pH 7.5 | Stable up to 6 months in IDTE pH 7.5 | |||
| Alt-R CRISPR enhancing reagents | HDR Enhancer* | Stable up to 1 year stored at stock concentration | Stable up to 1 year stored at stock concentration | Stable up to 1 year stored at stock concentration | Stable up to 1 year stored at stock concentration |
* The HDR Enhancer must be stored in a sealed container at –20°C. Under these conditions, the product should be stable for one year after the date of manufacturing (DOM). Purchasers are solely responsible for all decisions regarding the storage and use of these products and their suitability for the field of use, manner, and application for which such products are intended. Purchasers are also solely responsible for any associated legal or regulatory obligations.
** Data in this table represents unopened shelf life. Empty boxes indicate no data. ATTO is a trademark of ATTO-TEC, GmbH. Download a PDF of the table.
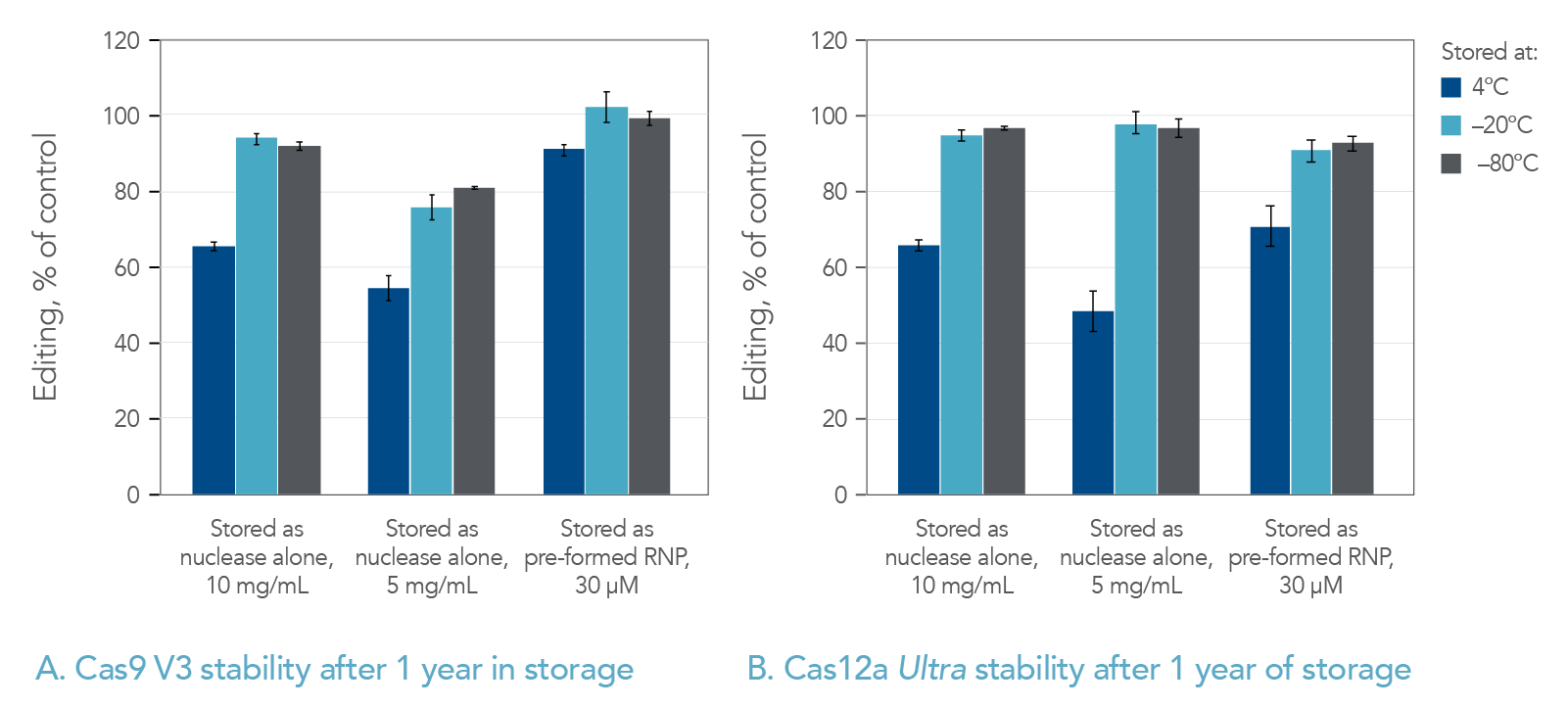
Nuclease and RNP stability
CRISPR nucleases complex with target-specific guide RNAs to form activated ribonucleoprotein (RNP) complexes. In practice, when working with Cas9, this is accomplished either by adding a single guide RNA (sgRNA) or an annealed 2-part guide RNA (containing crRNA and tracrRNA) to the Cas9 enzyme. When working with Cas12a, a single crRNA is added to the nuclease. Stability studies of both Cas9 and Cas12a RNPs were conducted. Storage of pre-complexed RNP showed no loss in storage stability in comparison to non-complexed nucleases at –20 and –80°C, over one year’s time (Figure 1). At 4°C, there was no loss of activity over 2 months (Table 1 and data not shown), but after a year at 4°C there was a partial loss in activity (Figure 1).
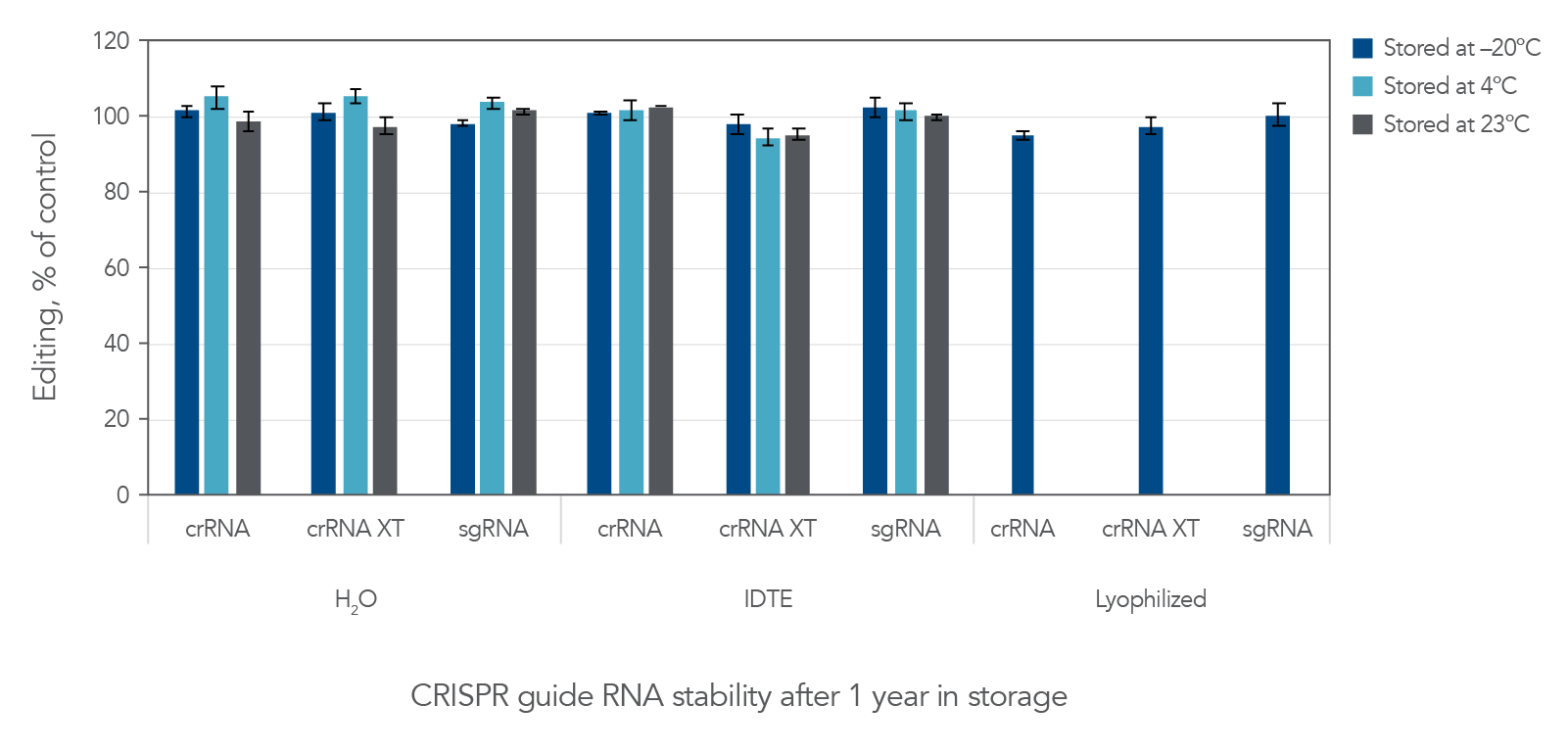
Effects of storage buffers and storage formats on guide RNA stability
As shown above in Table 1, we tested the stability of the Alt-R guide RNAs in both IDTE buffer and in nuclease-free water and compared these conditions to storage of lyophilized guide RNAs. RNA can be degraded in the presence of RNases found ubiquitously on human skin and in lab environments. However, when handled properly, the Alt-R guide RNAs were all stable in water (nuclease-free) as well as in IDTE buffer (Figure 2 and Table 1). They were also stable in lyophilized format at –20°C. Similar results were obtained with Alt-R tracrRNA stored in these conditions (data not shown). If users choose not to hydrate gRNAs upon receipt, it is important to store lyophilized samples at –20°C.
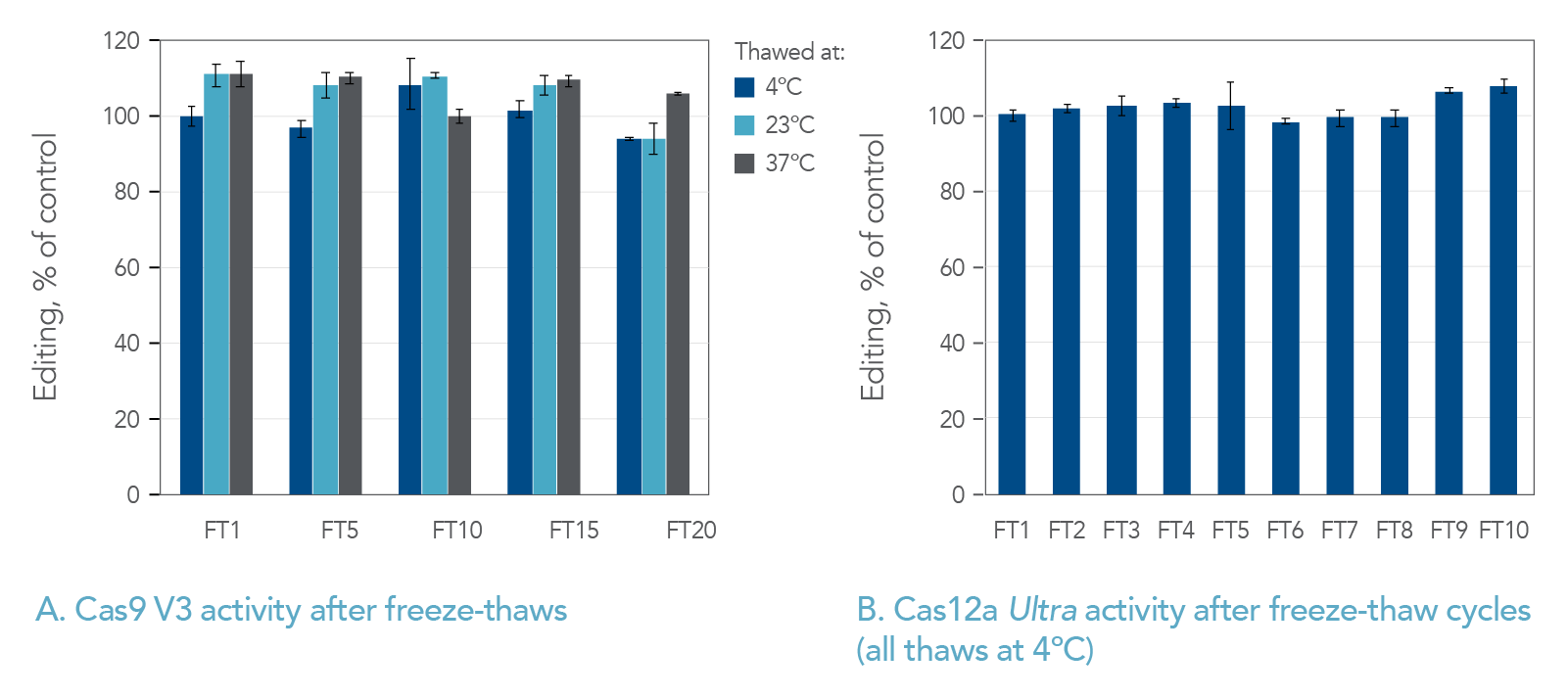
Effects of different thawing temperatures and increasing numbers of freeze-thaw cycles on Alt-R CRISPR nucleases
We investigated whether thawing Alt-R CRISPR reagents at different temperatures affects their stability after one or many cycles of freezing and thawing. We specifically tested conditions that would be easily replicated in most users’ laboratories. Starting with liquid solutions, we put the samples directly into a –80°C freezer. After the samples had frozen completely solid, they were placed either into a 4°C refrigerator to thaw, or on the bench at room temperature (23°C), or in a 37°C temperature-controlled metal heat block. We tested Alt-R Cas9 V3 through 20 freeze-thaw cycles at these multiple temperatures (Figure 3A), and we tested Cas12a Ultra and HiFi Cas9 through 10 freeze-thaw cycles at 4°C (Figure 3B, data not shown). We found that all the Alt-R CRISPR nucleases maintained complete functionality after many freeze-thaw cycles.
Conclusion
Although many biomolecules are greatly affected by freeze-thaw cycles and storage at different temperatures, Alt-R CRISPR reagents are remarkably stable in a wide range of conditions commonly encountered in research laboratories. Users can confidently store these reagents at the recommended temperatures and subject the reagents to repeated freeze-thaw cycles as shown in the above data without worrying about loss of activity.

