What is CRISPR screening?
CRISPR screening is a large-scale genetic loss-of-function experimental approach designed to find the equivalent of a few needles in a haystack. CRISPR screening facilitates discovery of key genes or genetic sequences that elicit a specific function or phenotype for a cell type (for a few examples, see Table 1). Like all good scientific experiments, CRISPR screening experiments are designed with a hypothesis in mind, but unlike many CRISPR experiments, the hypothesis is not a narrow one. All CRISPR screening experiments have the broad hypothesis that there are a few genetic sequences or genes in the genome that have a certain physiological effect, and that these few genetic sequences can be identified.
The result of a successful experiment is a short list of candidate genes or genetic sequences that appear to participate in producing the physiological effect under investigation. Therefore, CRISPR screening experiments not only start with a broad hypothesis, they end by generating new, narrow hypotheses. That is, each identified gene or genetic sequence usually needs to be investigated further using other biological methods to determine if it really produces the effect being studied.
CRISPR, as it is used by many researchers, is a method of making double-strand cuts at specifically targeted sites in DNA. When such cuts are produced in genomic DNA in cells, the cells use their DNA repair systems to mend the cuts. Commonly, the repair process is imprecise, and it results in mutations that knock out the targeted gene. This knockout event is the main result that scientists want for most CRISPR screening experiments [1]. CRISPR is described in much more detail on our website.
The CRISPR Basics Handbook from IDT
Download The CRISPR Basics Handbook, covering applications of CRISPR technology from guide RNA design to data analysis.
Table 1. Examples of uses of CRISPR screening
| Goal | Reference |
|---|---|
| Identify genes or DNA sequences causing cells to be either resistant or sensitive to a drug | [2] |
| Identify genes or DNA sequences affecting susceptibility to environmental toxins | [3] |
| Identify components of a cellular pathway | [4] |
| Identify genes or DNA sequences leading to a particular disease state | [5] |
How does CRISPR screening work?
Most CRISPR screening is done in cell culture. A few papers have described CRISPR screening in animals, and this will be described in more detail below. However, the main idea is easier to understand in cells, so we’ll start our description here.
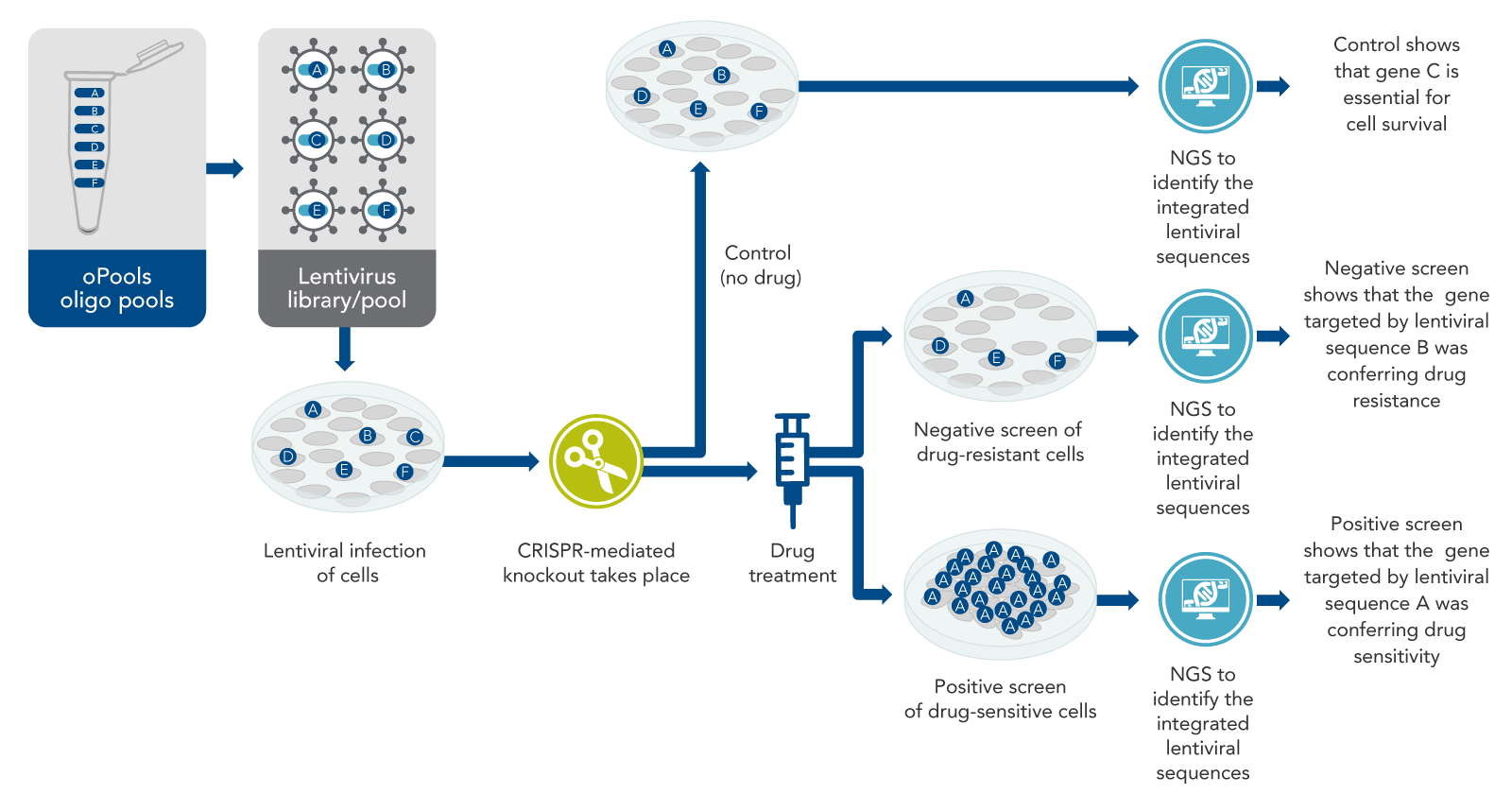
The basic idea of CRISPR screening is to knock out every gene that could be important, although knock out only one gene per cell (Figure 1). The intended result is a mixed population of cells with a different gene knocked out in each cell. In pooled screening experiments, this mixed population of cells with various genetically encoded perturbations are pooled together in the same dish. Some cells will die, but others will survive, or even excel in growth, becoming the predominant cell type. After the knockout cells are allowed to grow for a few days, next-generation sequencing (NGS) is performed on the entire mixed population of cells to determine which sequences are present and which are depleted, or absent. Such an experiment identifies genetic sequences necessary for survival under normal conditions. However, another aim addressed in most CRISPR screening studies is the identification of genes that allow cells to survive under specific conditions, such as drug treatment or other physiological situations of interest.
Fundamentally, as the basis for most CRISPR screening experiments, there is a specific physiological situation that needs to be understood better. For example, a cancer cell line may be resistant to some drug. The drug kills other cells but not this cancer cell line. Scientists usually start with a list of thousands of genes or genetic sequences that might be involved in this drug resistance and aim to narrow this list down by CRISPR screening. Often, screens start with a list of all genes in the genome to make sure nothing is missed.
From this comprehensive list of genes or genetic sequences, scientists then generate a long list of CRISPR targets. These targets are ~20-base DNA sequences located in the genome adjacent to sites known as protospacer-adjacent motifs (PAMs). For CRISPR screening, it is essential to knock out all the genes being studied. So, to increase the probability of cutting, several target sites must be selected for every gene or genetic sequence being studied. For pooled, vector-based screens, approximately 6–8 target sites per gene are recommended [6], although some researchers have had success targeting fewer sites per gene than this [7].
Control sequences: A properly designed CRISPR screening experiment should have numerous control sequences. These include the following:
- Negative controls: Negative control sequences should be designed not to have genetically- mediated physiological effects, so they can help identify any indirect confounding effects from the experimental materials and procedures. For genome-wide screens, as few as 100 such control sequences have successfully been used [8]; however, other authors recommend using around 1000 control sequences [9]. Negative control sequences often include the following:
- Non-targeting DNA sequences [10]. These non-targeting control sequences can either be designed without any homology to the cell’s genome, or they can have homology to a sequence that does not have an identifiable PAM nearby [11]. These controls should not cause cutting of the DNA and therefore should not have physiological effects.
- Controls that target regions of the genome not known to contain any genes [1]. These “safe-targeting” control sequences may or may not truly be “safe,” as they may cause unexpected biological effects.
- Sequences targeting genes already known not to have any effect on the physiological response under study. This is another kind of “safe-targeting” control [11].
- Positive controls: These are highly recommended when available. For this, it is necessary to know something about the biological effect being studied [11]. That is, if a specific gene is known to be involved in the biological effect, the pool should include several sequences targeting that gene.
Once the target and control sequences are identified, the next step is usually to design a pool of oligos which will be used to make lentiviruses. Other systems, such as adeno-associated virus, may also be used [12,13]. Another approach, is arrayed screens, which uses gRNA targets arrayed in a plate format as opposed to DNA oligo pools, and is described near the end of this article.
For the commonly used pool approach, each oligo in the pool must contain DNA to encode at least the targeting region of the CRISPR guide, or frequently the entire single-guide RNA (sgRNA) including the target sequence [7]. Each oligo also must have sites at each end to allow cloning into lentiviral gene-containing plasmids appropriately designed for biosafety [9,14]. From the plasmids, lentiviruses are produced as a pool containing thousands of CRISPR-targeting sequences, with one targeting sequence per virus particle (virion).
What is a CRISPR library and how is it used?
The term “CRISPR library” is often used interchangeably with terms such as “CRISPR guide RNA library” or “sgRNA library.” Traditionally, this “RNA library” is really the batch of lentiviruses produced from the pool of oligos. Considering this, such a library is not actually free RNA, even when it is called an “RNA library.” (More recently, synthetic gRNA CRISPR libraries have become popular, as described at the end of this article.)
Because it is true that lentiviruses are RNA viruses, each lentivirus in the library contains RNA, but this is not CRISPR guide RNA—this is viral RNA—and is much too long for a Cas enzyme to use in genome editing. When lentiviral libraries infect cells, CRISPR does not begin immediately since there is no CRISPR guide RNA yet. Instead, lentiviral RNA is first reverse-transcribed to DNA, which in turn is integrated into the genome of infected cells [7]. This integration can occur at any of several hundred sites in the cellular genome [15]. Even though these sites are not related to the CRISPR target sites, this integration may still affect the function of the genes surrounding the insertion site and ultimately impact the results of the experiment.
CRISPR lentiviral screening
In CRISPR lentiviral screening experiments, scientists aim to infect the cells with no more than one virion per cell. To make this practical, a very small ratio of virions to cells, or multiplicity of infection (MOI), is often used in the range of 5–30% [16]. Because each lentivirus in the library includes one sequence from the oligo pool, one such sequence is integrated as DNA into the genome of each of the infected cells. After integration, the lentiviral sequences—including the cloned-in CRISPR sequences—are transcribed to RNA. Thus, CRISPR guide RNA is eventually produced after the library of lentiviruses is used to infect cells [7].
The Cas enzyme
It is necessary for the cells to express a Cas (CRISPR-associated) enzyme for CRISPR screening to work. The CRISPR guide RNA targets the Cas protein to the intended site. Then, the Cas enzyme cuts the DNA. There are several methods that scientists use to deliver a Cas enzyme into cells for lentiviral CRISPR screening, including:
- Using a cell line that stably expresses a Cas enzyme. Such a cell line is often produced in advance using a separate lentivirus carrying the gene for the desired Cas enzyme, some time before starting the lentiviral CRISPR library screen [16].
- Producing a pool of lentiviruses that contain both the gene for the desired Cas enzyme, and the DNA that codes for the guide RNA [16].
Why are lentiviruses used in CRISPR screening?
Lentiviruses are potentially dangerous, and appropriate safety measures must be taken [14]. So, why are they so popular for CRISPR screening? As mentioned above, lentiviruses stably integrate their DNA into mammalian genomes [7,15]. This feature is used in CRISPR screening as follows. When cells that are infected with lentivirus survive, they undergo integration of the lentiviral genome, including the ~20 CRISPR targeting bases that have been cloned into the lentiviral genome. The cells continue to carry these guide sequences and express the corresponding CRISPR RNA (crRNA), even after the genes of interest are knocked out. When the cells proliferate, successive generations of cells carry the lentiviral DNA coding for guide RNA. This is useful because DNA or RNA from the cells can be sequenced, so scientists can discover which DNA/RNA sequence each cell contains. The continued expression of this DNA or RNA (after some cells have had a chance to proliferate, while other cells have been killed) is exactly what is measured in the CRISPR screen.
How many cells are needed?
As mentioned, an MOI of under 30% is usually used to try to ensure that no cell is infected with more than one virion (and thus more than one CRISPR targeting sequence). This means that many more cells are needed than just the ones that are infected. In addition, it is essential to infect at least 500 cells per targeting sequence to be sure of obtaining results within the sensitivity of the assay [9]. Therefore, for a CRISPR library that contains 100,000 targeting sequences, at least 1.67 x 108 cells are recommended [9]. Because the library is pooled, the cells are screened in large dishes, not in multi-well plates [6].
After infection, what happens?
After treatment of the cells with the viral library of targeting sequences and the desired Cas enzyme, the cells must be incubated for a few days. This allows the cells time to develop phenotypic CRISPR-mediated changes—in many experiments, this means the cells either grow or die off (Figure 1). Then, drug or other treatment is performed if desired for the specific experiment. After this, both total genomic DNA samples, or total RNA samples, from the two mixed populations (i.e., the control cells and the drug-treated cells) are collected for sequencing. The DNA (or RNA) is subjected to sequencing by NGS. The two resulting lists of sequences (control vs. drug-treated) can then be compared.
Negative vs. positive screens
Drug resistance and drug sensitivity are two of the major physiological responses that are frequently studied by CRISPR screening (Figure 1). Negative screens are used to find genes that cause drug resistance, and positive screens are used to find genes that cause drug sensitivity. These approaches are very similar, but the outcomes are different, as described below.
Negative Screens
Returning to the example of a cancer cell line that is resistant to a specific drug, how does screening work? If the resistant cancer cell line is first CRISPR-screened and then treated with the drug (which usually does not kill the cells, because they are resistant), some of those cancer cells will actually die in response to the drug. This suggests that resistance genes were knocked out. The control for this experiment would be cells targeted with the same lentiviral pool, but not treated with the drug. This is necessary because if essential genes are knocked out in some cells, those cells would die even without the drug.
There will be many sequences detected from no-drug control cells, because most of these cells survive. However, for the experimental sample (drug-treated cells), the target sequences that knocked out the resistance genes will not be detected by sequencing. The missing sequences have a high probability of having targeted the drug resistance genes. This is called a negative screen (Figure 1). To confirm the findings of a negative CRISPR screen, other kinds of biological studies usually are done.
Positive Screens
Cell types that are sensitive to a particular drug are naturally killed by the drug. To identify the genes that confer sensitivity, scientists can use the same method described for resistant cancer cell lines. In this case, some of the guide RNAs in the screen may knock out genes that cause sensitivity to the drug. The unedited cells (which are a majority of the cells after the CRISPR screen because the MOI is less than 30%) will be killed by the drug. Furthermore, most of the CRISPR-edited cells will also be killed because most genes in the genome are not drug-sensitivity genes. Only a few cells—expressly those with the drug-sensitivity genes knocked out—will grow.
Cas enzyme
These cells proliferate far more than the other cells in the presence of the drug and may take over the population. Sequencing by NGS will show the presence of the DNA encoding the guide RNA that was used to delete the sensitivity gene. This DNA will be present in large amounts, because the cells carrying this DNA have proliferated after their sensitivity gene has been knocked out. This is called a positive screen (Figure 1).
Why is it important to target multiple sites per gene?
Targeting multiple sites per gene is important for at least two reasons:
- To increase the probability of generating a cut within the targeted gene (as described earlier), as not all CRISPR targets are cut with equally high efficiency.
- To provide confidence of a real result. If only one sequence that targets a gene is missing (in a negative screen) or overrepresented (in a positive screen) in the final NGS analysis, this might be due to one of the following:
- Random chance or imperfect experimental setup
- Lentiviral integration into an important gene, thus knocking out that gene even before CRISPR takes place
- Off-target effects
However, if the results of multiple targets in the same gene are consistent, it is likely that these results are real. There is strong evidence that a gene responsible for the physiological effect has been identified [16].
More applications of CRISPR screening
In addition to looking for genes that cause drug resistance or drug sensitivity in cells, CRISPR screening has been used in many other contexts [17]. For example, it has been used to identify genes important in mitochondrial metabolism [4] and lysosome function [18]. With a catalytically dead Cas9 enzyme in conjunction with transcriptional activators, CRISPR screening has been used to turn on expression of thousands of long non-coding RNAs (lncRNAs) to identify those involved in regulating drug resistance in melanoma cells [19].
Other screening projects have used catalytically dead Cas9 both in combination with transcriptional activators to turn on expression of a large number of genes, as well as without transcriptional activators thus knocking down expression of many genes [20]. Another approach to CRISPR screening includes using two different targeting sequences per lentivirus within a library—an approach that has a variety of applications ranging from use of Cas9 nickases in screening to deletion of numerous large segments of genomic DNA in large screening projects [21].
CRISPR screening in animals
It is impractical for most labs to implement large CRISPR screens in animals, as this could require large numbers of animals each carrying different genetic mutations for a single experiment. However, some CRISPR screening methods have been used in animals. For example, in one project, a mouse cancer cell line which normally does not metastasize was treated with a CRISPR library of over 67,000 lentiviruses. The cancer cells were then transplanted into mice. Tumors grew and metastasized. By sequencing the DNA in the metastases, the researchers identified several CRISPR-targeted genes. This showed that loss of function of these genes may cause tumor growth and metastasis [5]. In another study, researchers used catalytically dead Cas9 fused to a cytidine deaminase base-editing protein system, which enzymatically alters the genomic DNA sequence by introducing G>A and C>T base substitutions without making double-strand breaks.
They treated mouse embryonic stem cells with an sgRNA library, using their base-editing system to insert specific mutations at 77 targeted sites. The mouse embryonic stem cells were injected into the cytoplasm of mouse oocytes, which were then implanted into female mice. The mice gave birth to offspring with the targeted mutations. This screen identified four amino acid positions in one protein essential for production of primordial germ cells. This novel screening technology provides a method for researchers to investigate protein sequence and function relationships in vivo [22].
CRISPR screening without a pool, using arrayed gRNAs
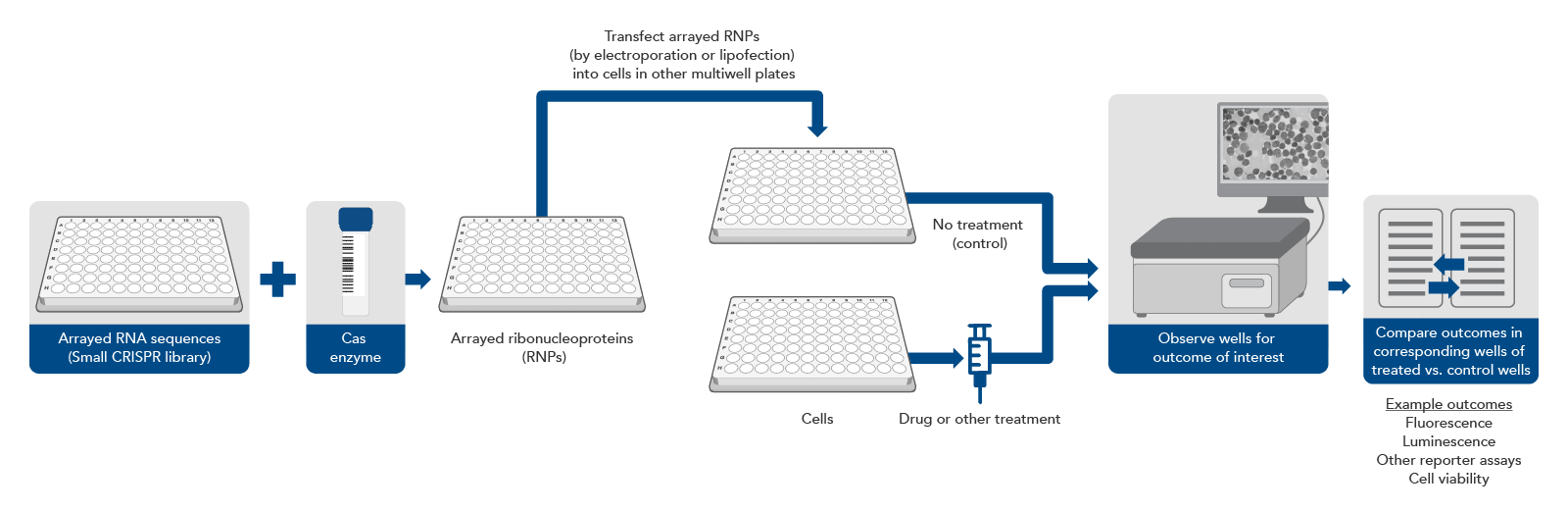
Traditionally, almost all CRISPR screening was done with a pool of lentiviruses carrying a large number of sequences to be targeted. However, scientists may already have a short list of genes of interest (for example, all the genes in one signaling pathway) and they want to verify a narrower set of targets that were identified in an initial lentiviral primary screen, or they may require a more sophisticated genotype-to-phenotype screening workflow. In either instance, a different approach can be taken (Figure 2).
A synthetic gRNA CRISPR library consists of synthetic RNA guides kept in separate wells in an array format on a multi-well plate. There are different formats for these arrays, but the most common are a single guide per well or multiple guides per gene, pooled together by gene, so that each well will knockout a single gene. In this experimental design, gRNAs can be complexed with an appropriate Cas enzyme in each well. These ribonucleoprotein complexes can be transfected, by electroporation or lipofection, individually into cells in different wells of a multi-well plate. Thus, each well of cells is transfected with one or several predetermined CRISPR targeting sequences. Then, depending on the experimental setup, the wells can be monitored for a physiological effect (live/dead cells, expression of fluorescent protein, or some other readout).
This approach eliminates the need for NGS but requires automation equipment to avoid being extremely labor intensive and can be much more expensive if a large library is used. However, by removing the need for lentivirus, as few as 2–3 target sites per gene need to be selected for high editing efficiency, as there is no chance that lentiviral integration will disrupt an important genomic sequence. However, even just one target site per gene can be sufficient if the target site is already well characterized and known to cause a knockout [11]. Avoiding lentivirus also removes biosafety requirements and assures that the synthetic gRNAs will not continue to exist in the cells after the knockout has occurred, decreasing the likelihood of off-target editing effects. Additionally, this approach is more appropriate for sophisticated screens where the parameters are around change in phenotype caused by the gene knockout, which can be viewed through microscopy, rather than just live or dead cells. This means that cell types that are non-dividing, such as primary cells and neurons, can also be screened since they do not lend themselves to the live or dead output with pooled CRISPR screens.
IDT offers custom CRISPR gRNA libraries. For more information, visit our Alt-R CRISPR gRNA libraries page, or contact CRISPR@idtdna.com.
Conclusion
CRISPR screens hold great promise for identifying genes and genetic sequences involved in many physiological pathways and pathological conditions. IDT offers custom synthetic gRNA CRISPR libraries for arrayed screens, or oPools oligo pools (pooled DNA oligonucleotides), to facilitate your production of libraries of lentiviruses carrying sequences encoding CRISPR guide RNAs.


 Processing
Processing

